
Eukaryotic DNA is packaged within the nucleus through its association with histone proteins (H2A, H2B, H3, and H4), forming the fundamental repeating unit of chromatin, the nucleosome. The precise architecture of chromatin dictates whether it is permissive or resistant to transcription and to other DNA-templated processes, such as replication, DNA repair, and recombination. While the majority of nucleosomes in a cell are composed of the same 4 types of core histones, tremendous diversity in the histone / nucleosome structures is generated by a variety of post-translational modifications, such as acetylation, phosphorylation, methylation, ubiquitinylation and sumoylation. These modifications occur on multiple but specific sites on the histones, and the different combinations of histone modifications may result in distinct outcomes in terms of chromatin-dependent functions such as gene expression, replication, or DNA repair.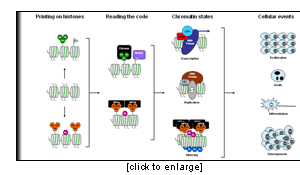
Our research – defined as a chromatinomics approach – focuses on the study of the functional network and the dynamics of the proteins that modify the histones and read these histone marks.







© 2006 - IRI Chromatinomics Group